Biological Science 2024/09/10
NAIST researchers explore previously unknown functions of Exportin-5 in exporting RNA molecules within Drosophila cells
Eukaryotic cells are complex biological units, each with multiple membrane-bound compartments. These cells use specialized mechanisms to export biomolecules from their synthesis site to where they function.
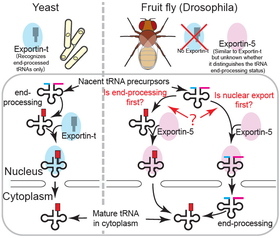
For example, messenger RNAs (mRNAs) are synthesized in the nucleus but must be exported to the cytoplasm for protein production. Specific proteins, such as the transcription-export complex, recognize features that are unique to mRNAs and manage their export. Other examples include transfer RNAs (tRNAs), which are recognized and exported by Exportin-t, and microRNAs (miRNAs), which are recognized and exported by Exportin-5 (Exp5).
Recent studies show that export mechanisms in fruit flies (Drosophila) differ slightly from other eukaryotes. In Drosophila, Exp5 also exports partially processed RNA molecules, including pre-tRNAs, indicating a broader range of functions than previously thought.
Aiming to close this knowledge gap, a team of researchers from Japan, led by Katsutomo Okamura from Nara Institute of Science and Technology, conducted a study to explore the role of Exp5 in Drosophila. Through their analysis of the RNAs associated with Exp5, they uncovered that Exp5 interacts with a broader spectrum of RNAs than previously recognized, including tRNAs, miRNAs, long non-coding RNAs (lncRNAs), and specific mRNAs. This research was published in the Journal of Biological Chemistry.
When asked about the motivation behind the study, Okamura says, "We aimed to explore the extent of evolutionary conservation of Exp5 functions by examining the substrate specificity of Drosophila Exp5."
To investigate the function of Exp5 in Drosophila, the researchers used the crosslinking and immunoprecipitation (CLIP) technique in Drosophila melanogaster-derived S2R+ cells. This method involved crosslinking RNA to the Exp5 protein with UV light and isolating the RNA-protein complexes to identify the RNAs bound by Exp5. The CLIP experiment successfully captured known Exp5 substrates, such as tRNAs and miRNAs and also identified new substrates, including various unique mRNAs and long non-coding RNAs (lncRNAs).
The study also highlighted that Exp5 might selectively bind and export specific RNA classes, particularly intronless mRNAs (transcribed from genes lacking non-coding regions called introns, which are typically "spliced out" or removed from the RNA transcript). The researchers arrived at these findings by comparing the RNA sequences recovered from the experiment with their expression levels. Okamura exclaims, "These results indicate that Drosophila Exp5 may have a broader range of substrates than previously believed."
The study also notably found that Exp5 attaches to immature tRNA molecules before the ends of these tRNAs are fully processed or trimmed. This finding indicates that Exp5 may play a role in the early stages of tRNA processing, helping to move these tRNA precursors out of the nucleus even before they are completely mature.
The researchers have, thus, proposed a new model explaining the function of Exp5 in RNA processing--Exp5 shuttles RNA molecules among different regions of the cell, ensuring the RNA molecules undergo proper processing rather than strictly selecting fully processed RNAs for export.
###
Resource
- Title: Exportin-5 binding precedes 5'- and 3'-end processing of tRNA precursors in Drosophila
- Authors: Ze Li, Junko Iida, Masami Shiimori, Katsutomo Okamura
- Journal: Journal of Biological Chemistry
- DOI: 10.1016/j.jbc.2024.107632
- Information about the RNA Molecular Medicine Laboratory can be found at the following website: https://bsw3.naist.jp/eng/courses/courses216.html